Computational Analysis
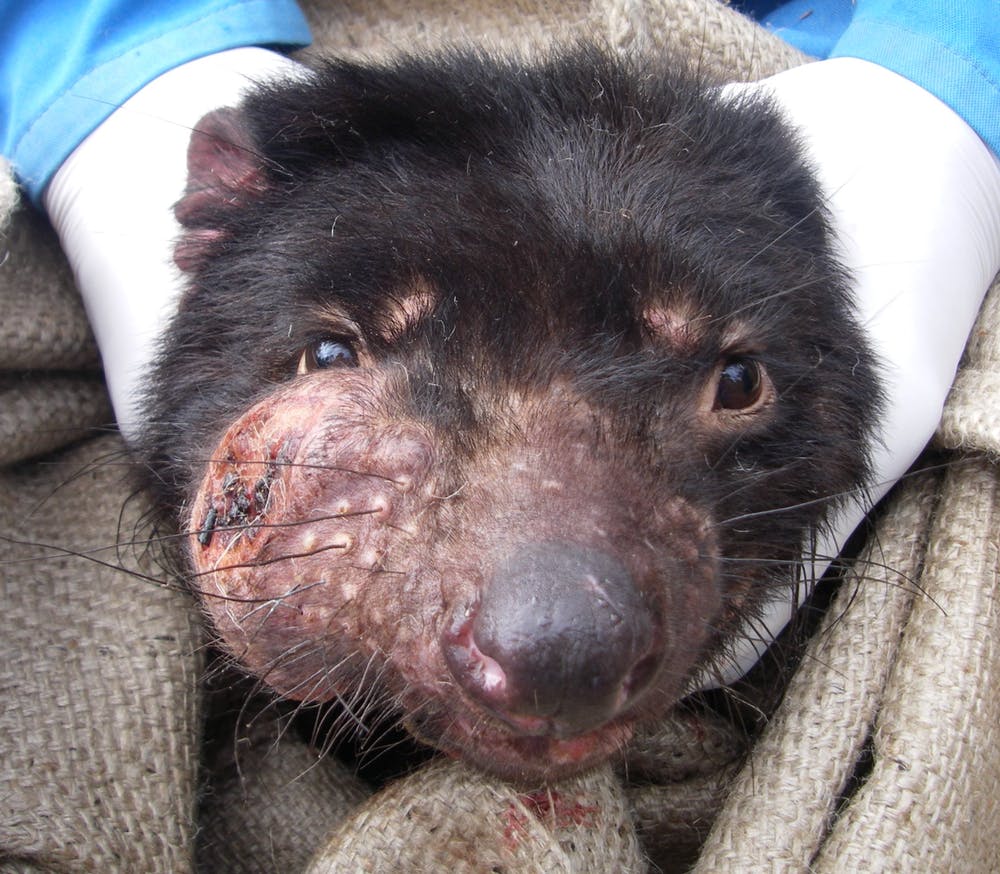
In high school, I completed a research paper using computational biology methods to compare the two known types of clonally transmissible cancer, Tasmanian devil facial tumor disease (DFTD) and canine transmissible veneral tumor (CTVT). To do this, I identified the specific protiens or genes that are considered to be markers for the different types of cancers and then employed several different biology databases such as Ensembl and UniProt to create computational profiles for the genes that represented both diseases. It was concluded that CTVT’s marker genes tend to be expressed in much more localized locations than DFTD’s, which is why DFTD is much harder to treat than CTVT.
Such a large undertaking was not taken alone. My professor, Robert Gotwals, offered assistance on an as-needed basis, and helped me figure out what databases were best to access and how to access them. He also taught me research and statistical methods that I utilized to come to my paper’s conclusions. I also received assistance from the North Carolina School of Science and Mathmatics, as they granted me access to many research journals that I perused in order to understand the necessary background for my paper.
Although this paper was a tremendous amount of work, I am ultimately glad that I went through such a process at such a relatively young age. It helped me narrow down potential career paths by introducting me to an interest in coding and steering me away from future research positions. I also honed my ability to research esoteric topics online, trawl through large databases, ask the right questions to get helpful answers, and be patient - such time-consuming papers and projects will never be completed in a day.